Primary structure of Protein
– In this topic, we will discuss The Primary structure of the Protein.
Primary structure of Protein
– The sequence of amino acids in a protein is called the primary structure of the protein.
– Understanding the primary structure of protein is important because many genetic diseases result in proteins with abnormal amino acid sequences, which cause improper folding and loss or impairment of normal function.
– If the primary structures of the normal and the mutated proteins are known, this information may be used to diagnose or study the disease.
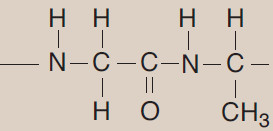
(A) Peptide bond
– In proteins, amino acids are joined covalently by peptide bonds, which are amide linkages between the α-carboxyl group of one amino acid and the α-amino group of another.
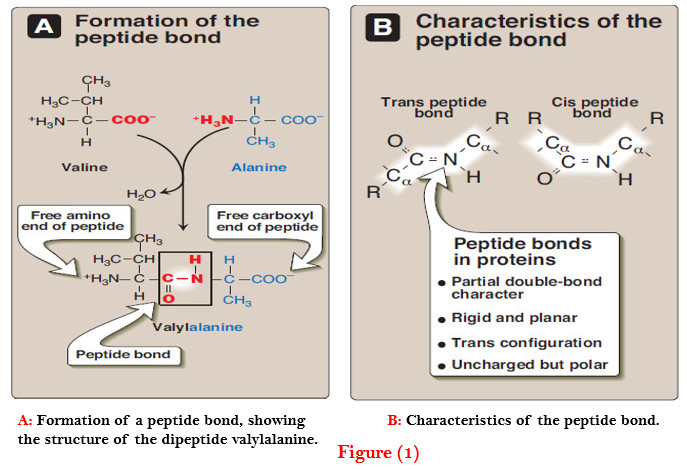
– For example, valine and alanine can form the dipeptide valylalanine through the formation of a peptide bond (Figure 1).
– Peptide bonds are not broken by conditions that denature proteins, such as heating or high concentrations of urea.
– Prolonged exposure to a strong acid or base at elevated temperatures is required to hydrolyze these bonds non enzymically
(1) Naming the peptide
– By convention, the free amino end (N-terminal) of the peptide chain is written to the left and the free carboxyl end (C-terminal) to the right. Therefore, all amino acid sequences are read from the N- to the C-terminal end of the peptide.
– For example, in Figure 1A, the order of the amino acids is “valine, alanine.”
– Linkage of many amino acids through peptide bonds results in an unbranched chain called a polypeptide.
– Each component amino acid in a polypeptide is called a “residue” because it is the portion of the amino acid remaining after the atoms of water are lost in the formation of the peptide bond.
– When a polypeptide is named, all amino acid residues have their suffixes (-ine, -an, -ic, or -ate) changed to -yl, with the exception of the C-terminal amino acid.
– For example, a tripeptide composed of an N-terminal valine, a glycine, and a C-terminal leucine is called valyl glycyl leucine.
(2) Characteristics of the peptide bond
– The peptide bond has a partial double-bond character, that is, it is shorter than a single bond, and is rigid and planar (Figure 1B).
– This prevents free rotation around the bond between the carbonyl carbon and the nitrogen of the peptide bond. However, the bonds between the α-carbons and the α-amino or α-carboxyl groups can be freely rotated (although they are limited by the size and character of the R-groups).
– This allows the polypeptide chain to assume a variety of possible configurations.
– The peptide bond is generally a trans bond (instead of cis, see Figure 1B), in large part because of steric interference of the R-groups when in the cis position.
(3) Polarity of the peptide bond
– Like all amide linkages, the – C=O and –NH groups of the peptide bond are uncharged, and neither accept nor release protons over the pH range of 2–12.
– Thus, the charged groups present in polypeptides consist solely of the N-terminal (α-amino) group, the C-terminal (α-carboxyl) group, and any ionized groups present in the side chains of the constituent amino acids.
– The – C=O and – NH groups of the peptide bond are polar, and are involved in hydrogen bonds, for example, in α-helices and β-sheet structures.
(B) Determination of the amino acid composition of a polypeptide
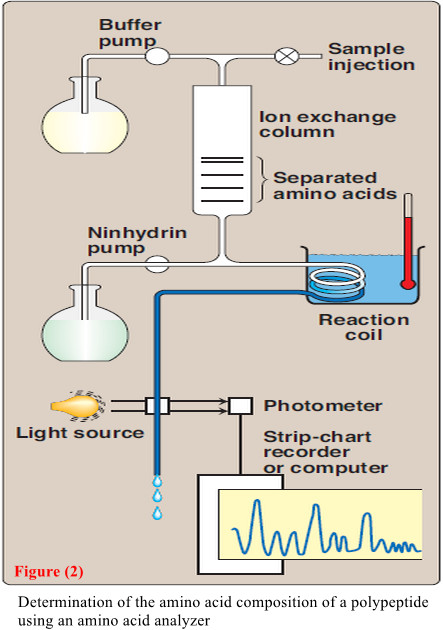
– The first step in determining the primary structure of a polypeptide is to identify and quantitate its constituent amino acids.
– A purified sample of the polypeptide to be analyzed is first hydrolyzed by strong acid at 110°C for 24 hours.
– This treatment cleaves the peptide bonds and releases the individual amino acids, which can be separated by cation-exchange chromatography.
– In this technique, a mixture of amino acids is applied to a column that contains a resin to which a negatively charged group is tightly attached.
– [Note: If the attached group is positively charged, the column becomes an anionexchange column.]
– The amino acids bind to the column with different affinities, depending on their charges, hydrophobicity, and other characteristics.
– Each amino acid is sequentially released from the chromatography column by eluting with sol utions of increasing ionic strength and pH (Figure 2).
– The separated amino acids contained in the eluate from the column are quantitated by heating them with ninhydrin—a reagent that forms a purple compound with most amino acids, ammonia, and amines.
– The amount of each amino acid is determined spectro photo metrically by measuring the amount of light absorbed by the ninhydrin derivative.
– also The analysis described above is performed using an amino acid analyzer—an automated machine whose components are depicted in Figure 2.
(C) Sequencing of the peptide from its N-terminal end
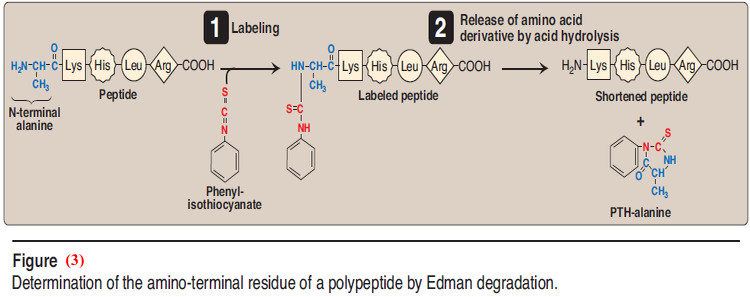
– Sequencing is a stepwise process of identifying the specific amino acid at each position in the peptide chain, beginning at the N- terminal end.
– Phenylisothiocyanate, known as Edman reagent, is used to label the amino-terminal residue under mildly alkaline conditions (Figure 2.4).
– The resulting phenylthiohydantoin (PTH) derivative introduces an instability in the N-terminal peptide bond that can be selectively hydrolyzed without cleaving the other peptide bonds.
– The identity of the amino acid derivative can then be determined.
– Edman reagent can be applied repeatedly to the shortened peptide obtained in each previous cycle.
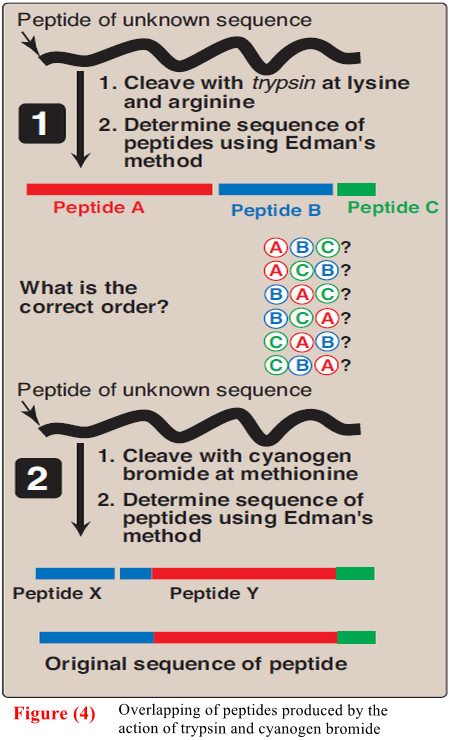
(D) Cleavage of the polypeptide into smaller fragments
– Many polypeptides have a primary structure composed of more than 100 amino acids.
– Such molecules cannot be sequenced directly from end to end. However, these large molecules can be cleaved at specific sites, and the resulting fragments sequenced.
– By using more than one cleaving agent (enzymes and/or chemicals) on separate samples of the purified polypeptide, overlapping fragments can be generated that permit the proper ordering of the sequenced fragments, thus providing a complete amino acid sequence of the large polypeptide (Figure 4).
– So, Enzymes that hydrolyze peptide bonds are termed peptidases (proteases).
– [Note: Exopeptidases cut at the ends of proteins, and are divided into aminopeptidases and carboxy peptidases. Carboxypeptidases are used in determining the C-terminal amino acid. Endopeptidases cleave within a protein.]
(E) Determination of a protein’s primary structure by DNA sequencing
– The sequence of nucleotides in a protein-coding region of the DNA specifies the amino acid sequence of a polypeptide.
– Therefore, if the nucleotide sequence can be determined, it is possible, from knowledge of the genetic code, to translate the sequence of nucleotides into the corresponding amino acid sequence of that polypeptide.
– This indirect process, although routinely used to obtain the amino acid sequences of proteins, has the limitations of not being able to predict the positions of disulfide bonds in the folded chain, and of not identifying any amino acids that are modified after their incorporation into the polypeptide (posttranslational modification).
– Therefore, direct protein sequencing is an extremely important tool for determining the true character of the primary sequence of many polypeptides.
References:
- Lehninger Principles of Biochemistry / David L. Nelson, Michael M. Cox/ 7th ed, 2017.
- Lippincott’s Illustrated Reviews: Biochemistry / Richard A. Harvey, Denise R. Ferrier/ 5th ed, 2011 / Lippincott Williams & Wilkins, USA.
- Harper’s Illustrated Biochemistry /Robert K. Murray, David A. Bender , Kathleen M. Botham / 28th, 2009/ McGraw-Hill Companies, Inc. USA.